MERFISH Mouse Brain Receptor Map
This dataset contains a MERFISH measurement of a gene panel containing 483 total genes including canonical brain cell type markers, GPCRs, and RTKs measured on 3 full coronal slices across 3 biological replicates.
To demonstrate the power of the MERSCOPE® Platform, we generated the MERFISH Mouse Brain Receptor Map that includes canonical cell type markers, nonsensory G-Protein coupled receptors (GPCRs), and Receptor Tyrosine Kinases (RTKs). We designed this specific gene panel because nonsensory GPCRs mediate signaling and may play vital roles behind brain ageing and neurodegenerative disorders. However, GPCRs are notoriously difficult to analyze. Thus, the ability to completely spatially profile GPCR expression across the brain with cellular context can assist with gaining a deeper understanding of brain tissue structure and function.
Three Ways to Explore the Spatial Dataset
1. MERSCOPE® Web Vizualizer
Using our interactive single-cell spatial visualizer tool, you can toggle between UMAP and Spatial view of the MERFISH Mouse Brain Receptor Map. Select a gene of interest from the drop-down list and click to add cells to highlight clusters.
2. Jupyter Notebook on Google Colab
Our Juypter Notebook loads and visualizes single-cell transcriptomics and imaging data from the Vizgen MERFISH Mouse Brain Receptor Map dataset including:
- DAPI staining across the full tissue slice
- Detected transcripts across the full tissue slice
- Detected transcripts, DAPI image, and cell boundaries within a single field of view
In addition, the Notebook performs single-cell analysis to cluster cell types and visualizes:
- UMAP and spatial single-cell views colored by the cell clusters
- Heatmap of the gene expression within each cell type cluster
A full tutorial is available that walks users through Vizgen's MERFISH Mouse Brain Receptor Map Jupyter Notebook and shows users how to execute the Notebook on Google Colab. The notebook sections can be run independently and the entire notebook takes about 40 minutes to run.
3. Download the Data
You can download the MERFISH data files directly to your computer to explore using your data analysis program of choice. Below is the list of downloadable file types:
- Information on all transcripts detected in the sample (List of Detected Transcripts)
- Single-cell information from cell segmentation (Cell by Gene Matrix)
- Mosaic images of the full tissue
Access Individual Data Files Below:
To cite this data, please use: Vizgen Data Release V1.0. May 2021
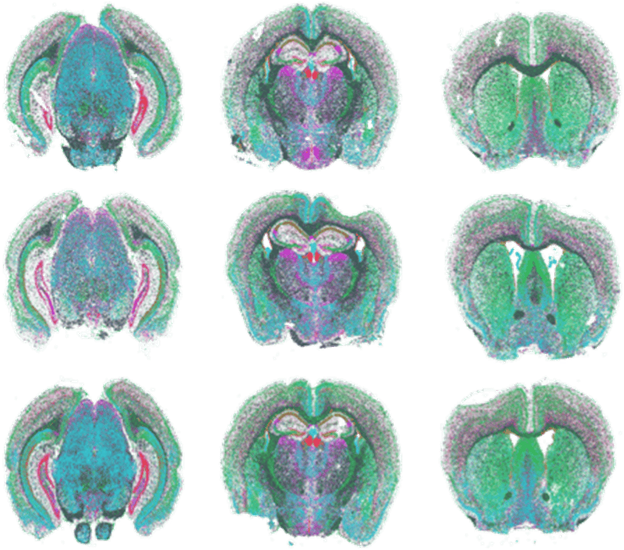
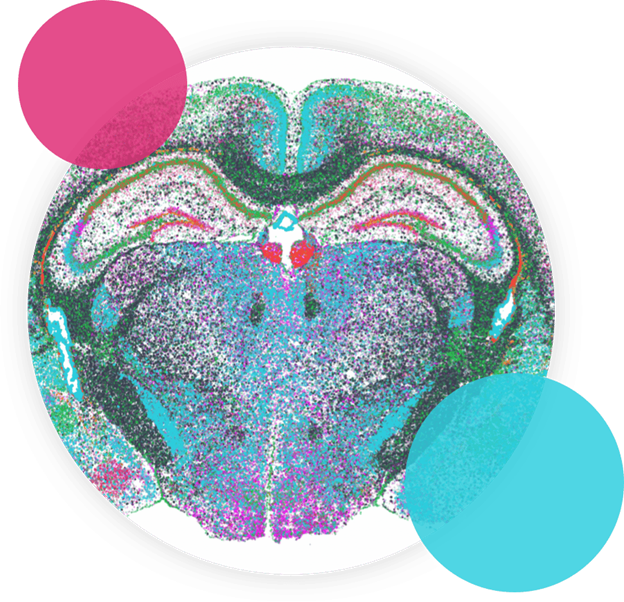
FIGURE 1
Figure 1: MERFISH measurements performed with MERSCOPE on nine mouse brain slices across three positions with three replicates per position.
Insights Gained from MERSCOPE Data
Detect 70X More Transcripts
Detect Lowly Expressing Genes With Subcellular Resolution
MERSCOPE detected 70x more transcripts per gene than an array-based platform for a more accurate map of the cellular organization of our selected mouse brain receptors:
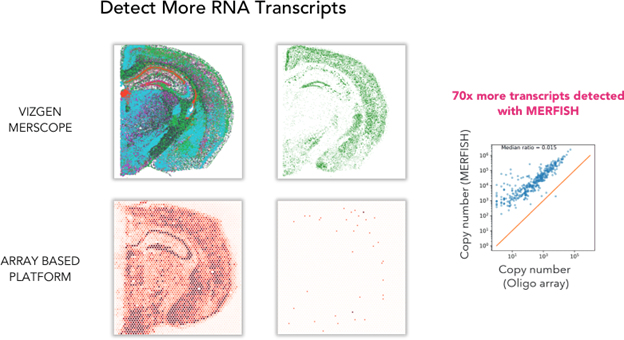
FIGURE 2
Figure 2: A comparison of total RNA detected between the Vizgen MERSCOPE and an array based spatial transcriptomics platform shows that MERSCOPE detects 70 times more transcripts from mouse brain tissue samples.
MERSCOPE is capable of a high 100nm resolution that enables us to see where RNA transcripts are located not only within a large 1cm2 tissue section, but inside each single cell, including Oxtr, Tshr, and Insr.
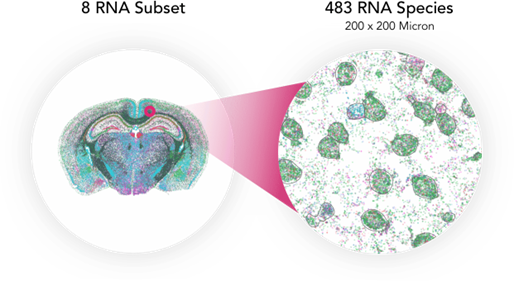
FIGURE 3
Figure 3: MERSCOPE can image a large 1cm2 area and provide the location of RNA transcripts within a tissue slice down to the subcellular level with 100nm resolution.
Map Gene Expression by Cell Type
Highly Reproducible MERFISH Measurements
MERSCOPE's high sensitivity allows us to map gene expression by cell type, as well as the variation in the gene expression pattern of receptors within the mouse brain.
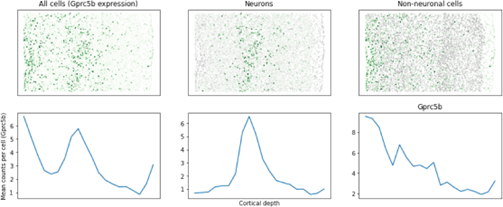
FIGURE 4
Figure 4: The variation in the gene expression pattern of eight different receptors between neuronal and non-neuronal cell types and their positions within the mouse brain are detectable by MERSCOPE.
MERSCOPE generates high reproducible measurements between brain slice replicates, resulting in high confidence in the data.
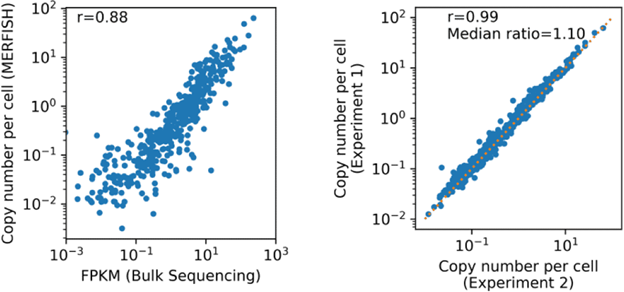
FIGURE 5
Figure 5: Correlation plots between the MERFISH measurements taken with MERSCOPE and bulk RNA-seq show that data generated by MERSOPE is highly quantitative and reproducible across large brain tissue area.
Vizgen MERFISH Brain Receptor Map
Our MERFISH Mouse Receptor Map demonstrates that the MERSCOPE Platform is a leading tool for building a superior molecular atlas because it produces true spatial transcriptomic data for whole tissue slices down to the subcellular level. MERSCOPE enables researchers to harness the power of MERFISH technology and gain greater insights into brain cell function.










